|
|||||
 |
|||||
Next: Results Up: Image Registration Previous: Transformation Parameterization
Phantom, CT, and MRI Image Data
The inverse consistency and transitivity error of the transformations produced using the unidirectional and consistent linear-elastic algorithms were evaluated using phantom, CT, and MRI image data (see Figs. 3, 4 and 5)2.
|
|
||||||
The phantom images were used to test how well the algorithms perform
using simple shapes and how well they perform when images contain large
regions of constant intensity. The dimensions of the phantoms were
![]() pixel images. Phantoms B and C were generated from image
A by deforming A with a random transformation such that the center of
mass of the images remained in the center of the image.
pixel images. Phantoms B and C were generated from image
A by deforming A with a random transformation such that the center of
mass of the images remained in the center of the image.
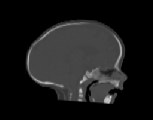
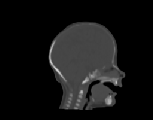
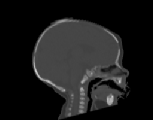
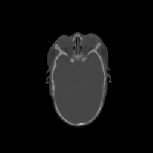
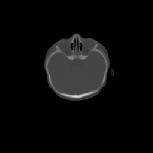
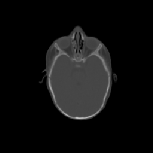
The CT images was used to test how well the algorithms perform when
registering CT data of the head. Specifically, we were interested in how
well the skull of one data set aligns with the skull of another data set
for pre-operative surgical planning and post-operative evaluation. The
CT images were collected from infants 3 months old and were selected such
that there was a large shape difference between the heads. The CT image
volumes A, B, and C were collected from a normal infant, an infant with
bicoronal synostosis, and an infant with sagittal synostosis, respectively.
The shape of the head in data set B is compressed front-to-back and the
shape of the head in data set C is compressed side-to-side and elongated
front-to-back. The CT data was resized and padded to make a
![]() voxels with voxel dimensions of 1 mm
voxels with voxel dimensions of 1 mm![]() . Each CT image volume was translated to align the sella turcica
to voxel location
. Each CT image volume was translated to align the sella turcica
to voxel location
![]() and rotated to align the Frankfort Horizontal plane of
the skull to the voxel lattice using the procedure described in [33]. The intensity range of the data
was reduced from 16-bits to 8-bits using a linear window that mapped the
CT units of 600-2200 to the range 0-255.
and rotated to align the Frankfort Horizontal plane of
the skull to the voxel lattice using the procedure described in [33]. The intensity range of the data
was reduced from 16-bits to 8-bits using a linear window that mapped the
CT units of 600-2200 to the range 0-255.
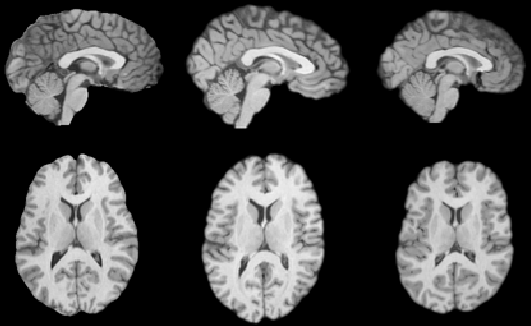
The MRI brain data was used to test how well the algorithms perform
on image volumes with complex-shape global and local anatomical structure.
Each MRI was manually edited to remove soft tissue, skull, and the spinal
cord. The MRI data sets were translated to align the anterior commissure
(AC) point with voxel location
![]() and rotated to align the midsagittal plane and the transverse
plane containing the anterior and posterior commissures with the voxel
lattice. The MRI data was resized and padded to create a
and rotated to align the midsagittal plane and the transverse
plane containing the anterior and posterior commissures with the voxel
lattice. The MRI data was resized and padded to create a
![]() voxel volume with voxel dimension 1 mm
voxel volume with voxel dimension 1 mm![]() . The intensity of the MRI data was converted from 16-bits to 8-bits
such that all of the MRI data sets had roughly the same shaped intensity
histograms.
. The intensity of the MRI data was converted from 16-bits to 8-bits
such that all of the MRI data sets had roughly the same shaped intensity
histograms.
Next: Results Up: Image Registration Previous: Transformation Parameterization Gary E. Christensen 2002-07-04
Copyright © 2002 • The University of Iowa. All rights reserved.
Iowa City, Iowa 52242
Questions or Comments: gary-christensen@uiowa.edu