|
|||||
 |
|||||
Next: Inverse Consistency Error Up: index Previous: Phantom, CT, and MRI
Results
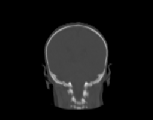
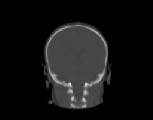
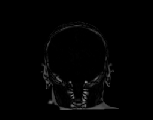
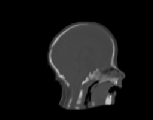
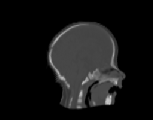
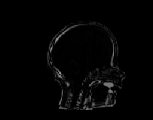
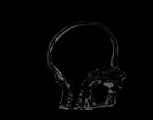
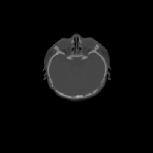
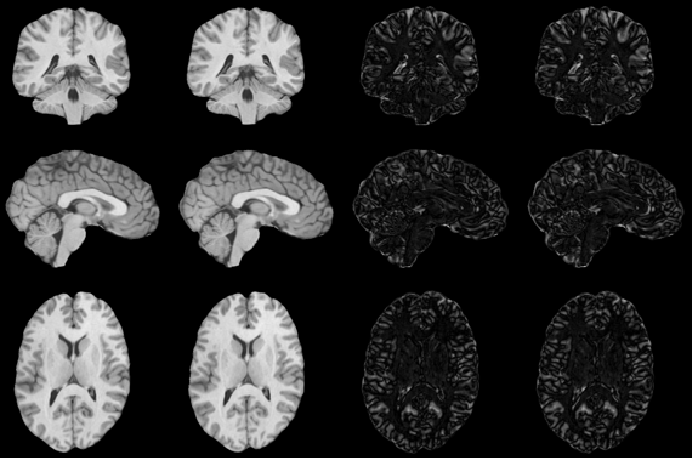
The inverse consistency and transitivity analysis was performed on three 2D phantom, three 3D CT, and 23 3D MRI data sets. The forward and reverse transformations between each pair of images were estimated using the unidirectional and consistent linear-elastic registration algorithms for each image modality. The image registration protocol used to register the phantom, CT and MRI images is described in Appendix A. All data sets were rigidly registered before being elastically registered. Typical transformation results for the phantom, CT and MRI data sets are presented in Figs. 6, 7 and 8. The CT and MRI figures show coronal, midsagittal, and transverse image slices from the 3D A-to-B registrations for the CT and MRI image registration experiments. These images represent the typical performance of the unidirectional and consistent linear-elastic image registration algorithms. Both figures show that the unidirectional and consistent algorithms produce very similar results with respect to matching image intensity. The difference images show that most of the matching error occurs at the edges of the images. This registration error is partially due to the fact that the linear-elastic model can only accommodate global nonrigid shape differences. The misregistration at the edges is also due to the balancing of the similarity and the regularization terms in the minimization problems defined by Eqs. 5 and 7. This occurs in both the unidirectional and consistent linear-elastic algorithms because the regularization term prevents the template image from fully deforming into the target data.
|
||||||||||||||||||||
|
|
||||||||||||
Subsections
Next: Inverse Consistency Error Up: index Previous: Phantom, CT, and MRI Gary E. Christensen 2002-07-04
Copyright © 2002 • The University of Iowa. All rights reserved.
Iowa City, Iowa 52242
Questions or Comments: gary-christensen@uiowa.edu
![\includegraphics[width=14cm]{trans_paper01.figs/phantoms/phantom_withouticc_1x4}](img90.png)